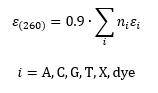Back to top
Frequently Asked Questions - Oligonucleotide Synthesis
Product & Services
The modification (3’ MGB-Q530) can be ordered directly through our webshop.
Yes, Microsynth offers Oligos containing degenerated Bases as a standard product. Degenerated Bases can be entered in the webshop directly using the respective IUB Code (See also Webshop). All degenerated Bases (mixed bases) are synthesized using highly accurate premixed synthesis reagents which ensure best possible equimolar distribution of the degenerated bases in the Oligo.
No, we recommend to use Primer-BLAST to design your primers.
Yes.
Microsynth primers can be further used in EvaGreen or probe based assays (e.g. drop-off assays for mutation screening).
The thiol-modified oligonucleotides purchased from Microsynth are already reduced to their active form. You can dissolve the thiol-modified oligonucleotides in the desired solvent and use them directly. Once dissolved, we strongly recommend storing the samples at -20°C or lower.
Over time, after being exposed to air or a solvent that could act as an oxidizing agent, such as DMSO, the reversible formation of an intramolecular disulfide bond may occur. In this case, the thiol-modified oligonucleotides need to be handled with a reducing agent like DTT or TCEP.
Both combinations are possible. However, based on our experience, HEX performs better with BHQ-2 than with BHQ-1. Therefore, we recommend using HEX in combination with BHQ-2 for optimal results.
Order Related Questions
Yes, we can deliver oligos in 384 well plates. Please download our excel order sheet under Upload entry in the webshop. Paste your sequences in the file and send the excel file by email to oligo.support@microsynth.ch. Do not submit the excel sheet directly in the webshop. We will come back to you with an offer and more details about your order.
Replace the different DNA/RNA or 2’-O-Methyl-RNA Bases in your sequence by the internal modifications 5, 6, 7 and 8 (e.g. TTAGCrAArGTTrUrC -> TTAGC5A6TT78). Login our webshop and enter your sequence into the sequence field. Define the modification in the drop down menu for 5, 6, 7, 8 (either RNA/DNA or 2’-O-Methyl-RNA bases).
You can shift the position of oligonucleotides in the 96 well plate by placing empty position. To accomplish that enter an oligo dataset with the following properties:
i. Oligoname: Leerposition
ii. Scale: Genomics
iii. Purification: Desalted
iv. Sequence: TT
The minimum amount of oligos to receive an 96 well plate is 40 oligonucleotides.
- Select a “T” DNA nucleotide within your sequence and replace the “T“ by “5”
- Select your preferred dye (e.g. FAM-dT) in the drop-down menu under “Inner Modification (5=...)”
- Follow the further instructions and submit your order
Initial screening using different siRNAs: for such an application our customers usually use desalted siRNA which has undergone cartridge purification. Herewith purity levels of ~80 % are achieved which is fairly enough for initial screening purposes.
Advanced screening or for use in cell cultures: if your experiment becomes more sophisticated and hence more expensive, we usually recommend to select HPLC (~85%) or even PAGE purification (~95%).
For larger quantities, starting from 5 mg, we do offer low endotoxin siRNAs, specifically for use in animal experiments. Please contact us for more information.
By standard, there is a OH-group at the 5’ and 3’ of an oligo.
Molar extinction coefficients of oligonucleotides are calculated according to the „base composition model“. [1-3] It includes the absorption of the individual nucleobases and potential base modifications and/or dyes. More accurate values for unmodified oligonucleotides are obtained from the „nearest-neighbor model“, which considers stacking effects of adjacent nucleobases. According to our calculations this model reduces the extinction coefficient approximately to 90 % compared to the base composition model. Therefore the extinction coefficients of oligonucleotides are estimated according to the below equation.
- Tataurov A.V., You Y., and Owczarzy R. (2008) Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids, Biophys. Chem. 133, 66-70.
- Fasman, G.D. (Ed.) (1975) Handbook of Biochemistry and Molecular Biology, Volume 1: Nucleic Acids, pp 589, 3rd edition, CRC Press.
- Cavaluzzi M.J., and Borer P.N. (2004) Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA, Nucleic Acids Res. 32, e13.