
Back to top
siRNA for Gene Silencing

Features and Benefits
- All siRNAs are MALDI-TOF MS controlled before shipment.
- Microsynth is a renowned siRNA supplier for top research institutes, pharma/biotech companies.
- Ready to use: siRNA is manufactured as ready to use aqueous solution (40 µM siRNA duplex, 20 mM NaCl, 10 mM TrisHCl, pH 7.5)
- An easy-to-use siRNA design tool for custom siRNAs is available in our webshop.
- Very competitive pricing
- High guaranteed yields
-
Our highly experienced and well-trained scientists are happy to support you.
- An easy-to-use online portal, a user-friendly input mask and other helpful tools (e.g. order tracking and history, convenient search and re-order option)
Overview
siRNA (small interfering ribonucleic acid) is a type of RNA that is involved in a number of biological processes, most notably RNA interference. RNA interference is a regulatory process that is used to control and limit the expression of specific genes. While most RNA is single stranded, siRNA is made up of two complementary strands of RNA nucleotides similar to those in DNA. siRNA molecules are usually 21-25 nucleotide double-stranded RNA (dsRNA) duplexes with symmetric 2 nucleotide 3’ overhangs.
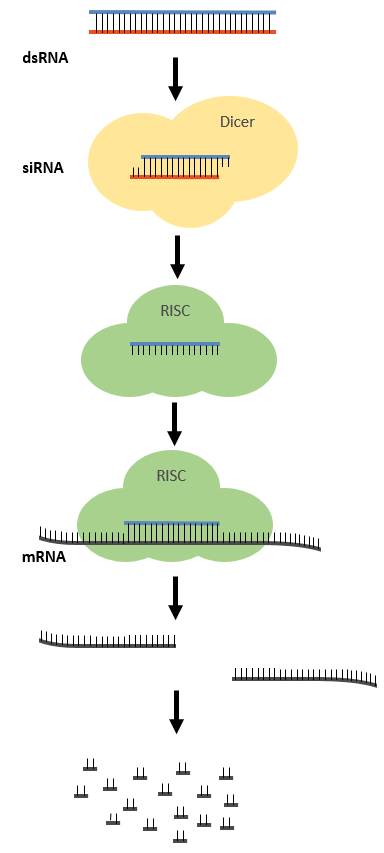
The RNA interference process and the biochemical machinery involved. Double-stranded RNA is cut into short pieces (siRNA) by the endonuclease dicer. The antisense strand is loaded into the RISC complex and links the complex to the mRNA strand by base-pairing. The RISC complex cuts the mRNA strand, and the mRNA is subsequently degraded.
non-modified siRNA Duplexes
Microsynth supplies siRNA duplexes 17-25 nt length, standardized ready to use in 40 uM (10 mM TRIS-HCl pH 7.5 and 20 mM NaCl).
Product Characteristics:
|
Synthesis scale1
|
Desalted
|
HPLC
|
PAGE
|
|||
|
OD2602
|
[nmol]2
|
OD2602
|
[nmol]2
|
[OD260] 2
|
[nmol]2
|
|
|
Genomics
|
on request
|
7
|
not available
|
not available | ||
|
0.04 µmol
|
9
|
21
|
4
|
8
|
not available
|
|
|
0.2 µmol
|
16
|
35
|
7
|
16
|
2
|
5
|
| 1.0 µmol |
36
|
84
|
16
|
45
|
5
|
11
|
|
Higher Scales
|
on request
|
|||||
modified siRNA Duplexes
For drug discovery, modified siRNAs are used to enhance their stability and half-life within cells and the body. Microsynth offers the most commonly used chemically enhanced siRNAs, ready for use. These include 2’ OMe, 2’ F, PTO, DNA, and 3’ GalNAc modifications.
Product Characteristics:
|
Synthesis scale
|
Modified siRNA, 2' OMe, 2' F, PTO, desalted quality, Na+ Form
|
Modified siRNA, 2' OMe, 2' F, PTO, 3' GalNAc, HPLC purified
|
||
|
OD260
|
[nmol]
|
OD260
|
[nmol]
|
|
|
0.04 µmol
|
6.4 |
20
|
0.64 | 2 |
|
0.2 µmol
|
12.8
|
40 | 1.92 |
6
|
| 1.0 µmol | 28.8 | 90 | 6.4 | 20 |
Control siRNAs
Positive and negative control siRNA are an essential complement to any gene silencing experiment. Positive controls are validated siRNAs that are known to achieve high levels (>70%) of knockdown. A positive control should be used to optimize transfection and reconfirm high levels of delivery in each RNAi experiment.
Non-target universal negative controls are validated siRNAs designed to show there is no homology to any known mammalian gene. They are important for distinguishing sequence-specific silencing from non–specific effects in the RNAi experiment.
Another negative control strategy is to use scrambled siRNA that has the same nucleotide composition but does not have the same sequence as the test siRNA. However, it is often impossible to design a scrambled control that has no known target in the cells being used.In the following table you will find a selection of siRNA controls that have been validated in numerous scientific publications.
| Name | Sense Strand Sequence (5’ -3’) |
Sense Strand |
Anti-Sense Strand 3’ Overhang (5’ – 3’) |
| Lamin | CUGGACUUCCAGAAGAACA | dTdT | dTdT |
| GFP | GCAGCACGACUUCUUCAAG | dTdT | dTdT |
| Luciferase | CGUACGCGGAAUACUUCGA | dTdT | dTdT |
| Non-targeting negative control | AGGUAGUGUAAUCGCCUUG | dTdT | dTdT |
| Scrambled negative control | On request | ||
As with any specific target siRNA, control siRNAs can be readily ordered in different synthesis scales and purification levels from Microsynth’s webshop.
siRNA Design
To help you in designing the most appropriate siRNA that targets your gene of interest, Microsynth has developed a proprietary software, and made it available in its webshop.
Microsynth’s siRNA design tool is making use of the set of guidelines that were initially provided by Boese et al [1]. The output of a design is various siRNAs that are ranked according to the Reynolds scores [2].
The siRNA sequences with the highest scores can be selected and directly ordered online.
A link to NCBI BLAST is incorporated so you can properly manage the risk of potential off-target effects of the predicted siRNA.
References:
[1] Boese Q, Leake D, Reynolds A, et al. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 2005;392:73-96. doi:10.1016/S0076-6879(04)92005-8
[2] Reynolds, A., Leake, D., Boese, Q. et al. Rational siRNA design for RNA interference. Nat Biotechnol 22, 326–330 (2004). https://doi.org/10.1038/nbt936
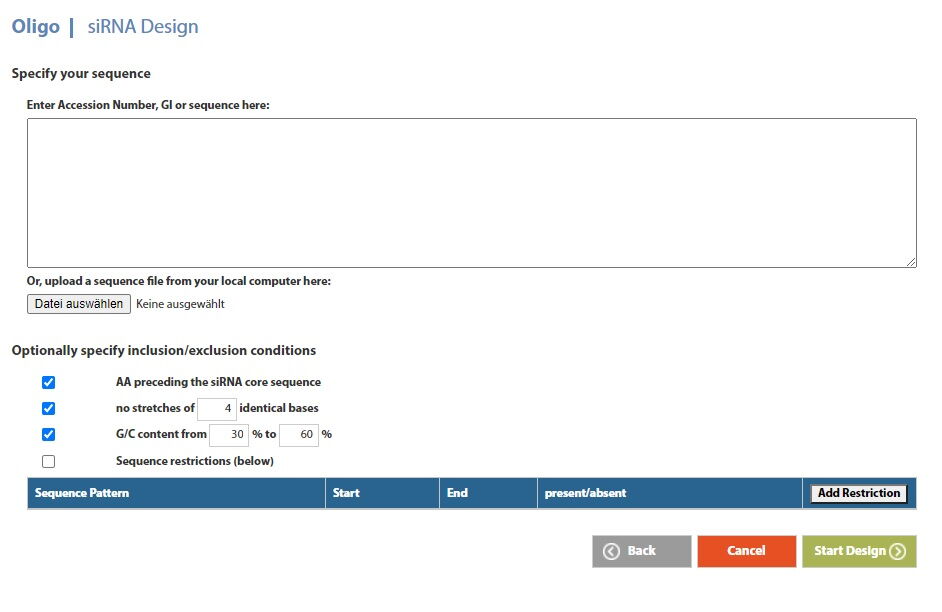
Figure: Microsynth's siRNA Design Tool
Specify your target sequence and, optionally, some additional parameters such as AA preceding the siRNA core sequence, the number of stretches of identical bases, the G/C content as well as further sequence restrictions (e.g. sequence areas that should be considered or not) to receive your design suggestion (highest scores on top) in a ready-to-order format.
How to Order
- Enter our webshop
- Click on siRNA in the blue “DNA/RNA Synthesis” domain
- Select Normal Entry in order to type or copy/paste the desired sequence information etc. or alternatively select Upload Entry by using our convenient Excel template (can be downloaded during ordering )

